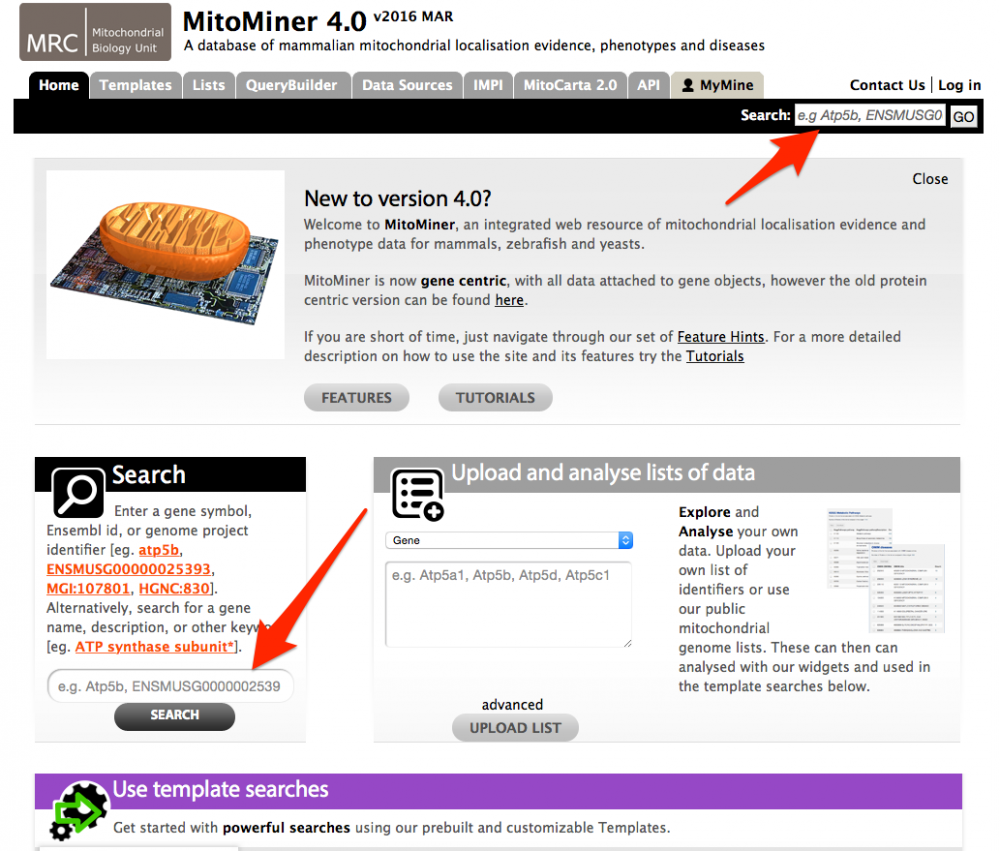
Tutorial 1. Quick Search
The Quick Search function provides a basic search facility for the data contained within MitoMiner and provides a good way to browse the database. The Quick Search function can be found on the front page and on the main menu bar, which is present on all pages. You can search for an ID or description for most parts of the database. Wild cards (*) can be added to increase the range of a quick search.
Some example search terms:
For Genes use gene symbols, Ensembl genes IDs, NCBI gene IDs, or genome project IDs (Hugo, MGI, RGD, ZFIN, SGD or PomBase)
For Proteins use UniProt identifiers, primary accessions numbers or descriptions (e.g. CY1_BOVIN or P16909 or Cytochrome c1).
For KEGG use KEGG identifiers or descriptions (e.g. 00010 or *glycolysis*).
For OMIM use a OMIM identifier or description (e.g. 125700 or *diabetes*).
For example to search for gene atp5b, the gene symbol of 'atp5b' would be entered into the search box and run by pressing the go button. A list of entries that match the search term are then displayed. For gene atp5b, four gene matches are displayed for four different species. Clicking on an entry will display its report page (described in the next tutorial).
You can also search for template queries (these are common queries that you can alter to your needs) and lists. This is done by selecting either the templates tab or the lists tab on the main menu bar. Then use the specific search box at the top of the page (for lists click on view link first).
To use Boolean terms such as AND, OR and NOT, be sure to include these in capitals. For example "Kinase AND human" will return records with Kinase and human in them. "Kinase and human" will look for all records with 'kinase', 'and' or 'human' in them.