Main menu
You are here
FAQ
Can I use my own list of genes?
Yes! These can be uploaded via the list uploader on the front page or on the 'Lists' tab. The 'Lists' tab also allows you to select the species - useful if you are uploading gene symbols. Your list can then be used in all relevant queries.
Why has an experimental dataset from <insert author here> not been included?
We are always on the look out for new datasets to be included in MitoMiner. If we have missed one that you would like included please contact us (MitoMiner at mrc-mbu.cam.ac.uk) with the publication details.
I can’t find a query that does....
New queries can be created using the query builder. See the tutorials for more details. Alternatively contact us with the details and we will be happy to help. If you have made a query that you think will be of use to other users, let us know and we can make it publically available.
How do I cite MitoMiner?
Please see the cite page.
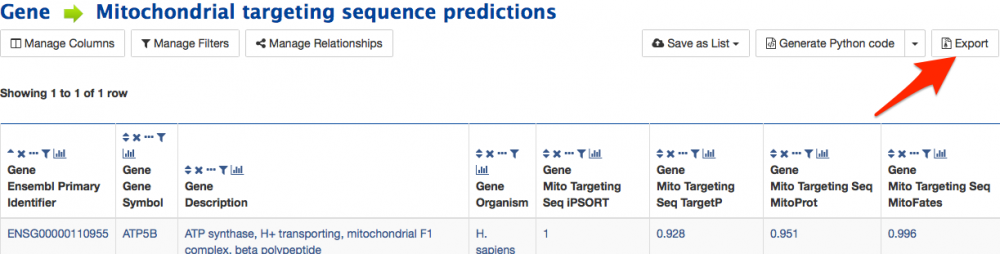
Once you have run a query it is possible to export the results using the export button on the header bar above the table (see screenshot below). The export button provides several options.
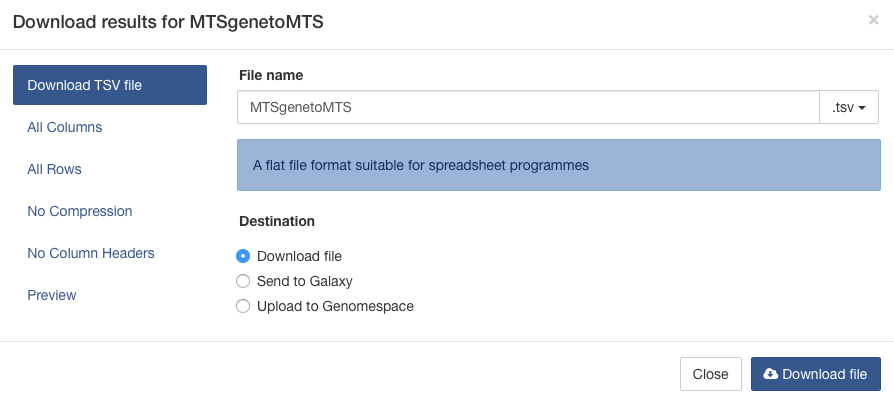
Select the format you would like such as TSV (suitable for MS Excel) to export the whole table. It is also possible to include additional columns in the results and reorder them by selecting the 'choose columns' button. To export a subset of the results select the 'Specific Range' button. Once you have made your selections simply click the green 'Download' button.