Main menu
You are here
Tutorial 2. Report page
Each entry within MitoMiner has a report page. This page serves as a summary of all the information present within the database that relates to that entry and provides web links to external resources such as the Ensembl and GeneCards databases.
All report pages have the same basic structure: The top part provides basic information for the entry itself, whereas the lower part shows information from other data sources that are relevant to this particular entry. The 'Quick Links' bar across the middle moves the page to the differect data source sections below. The 'External Links' section provides web links to external databases and the 'Lists' section shows any lists which have this entry including lists you have created yourself.
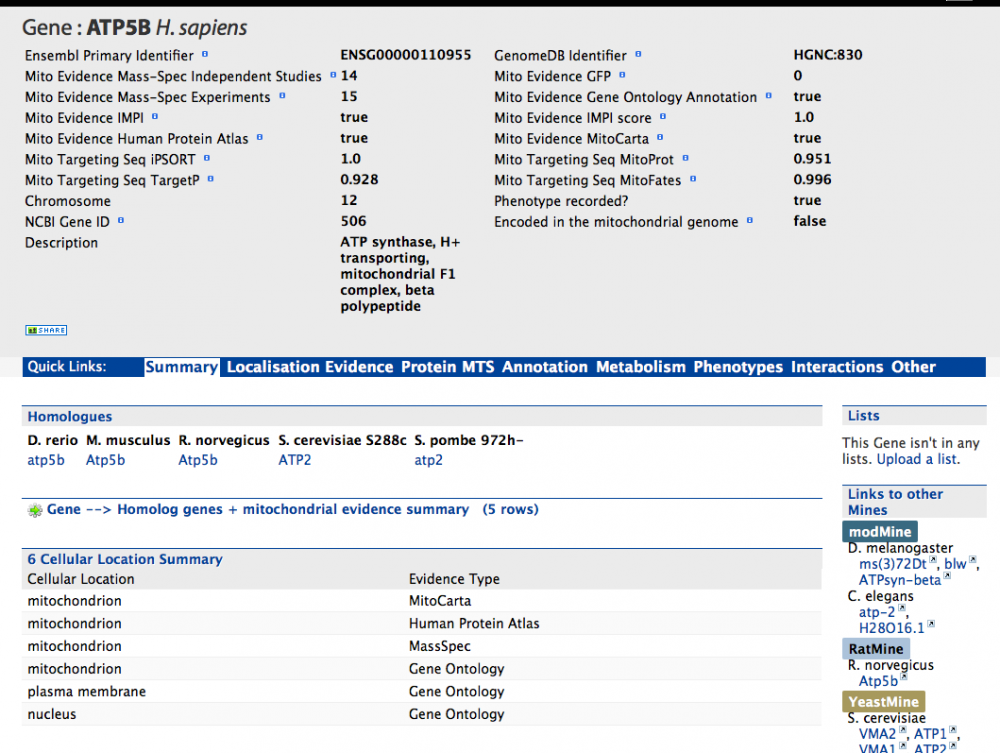
As an example the report page for gene ATP5B in human, is shown below. The summary at the top provides a brief overview of the gene. This includes basic information such as identifiers and its chromosome as well as various 'Mito Evidence' categories. These categories are gene specific and show a summary of the mitochondrial evidence for that gene, such as the number of mass-spec entries which have identified proteins encoded by this gene, whether it is annotated as mitochondrial in the Gene Ontology, and whether it is in the IMPI reference set of mitochondrial genes.
Below this summary is information from other data sources that are relevant (linked) to this particular gene. For example the 'Summary' sections shows homologs of the gene and the Gene Ontology annotation can be found in the 'Annotation' section.
The 'External Links' section in this case contains a web link to the relevant entry in the GeneCards database. Above that is the 'Lists' section, which shows this protein is in several lists such as the IMPI Reference set list. If you have created any of your own lists (see tutorial 5) a button will appear that will alllow you can add this particular entry to it here (as long as it is of the same type).